3. Project: Discover hidden diseases topics with LDA
Introduction#
Topic modeling for analyzing healthcare-related content has been used extensively. Latent Dirichlet Allocation(LDA) is one of the most famous topic modeling methods. LDA can help to discover hidden topics in document collections. In healthcare, it can be used for searching hidden disease topics.
For example, Prier et al.(2012) used LDA to identify several topics from health-related tweets, and found out that twitter users typically discussed strategies to quit smoking and recover from smoking addiction.
Our project#
In the healthcare analytics class, my team decided to use LDA and clustering to find hidden disease topics for patients who had depression. In another word, we would like to know what other diseases did the depressed patients also had. The data being used here is the CMS 2009 inpatient dataset. After preprocessed, let’s have a preview of the data:
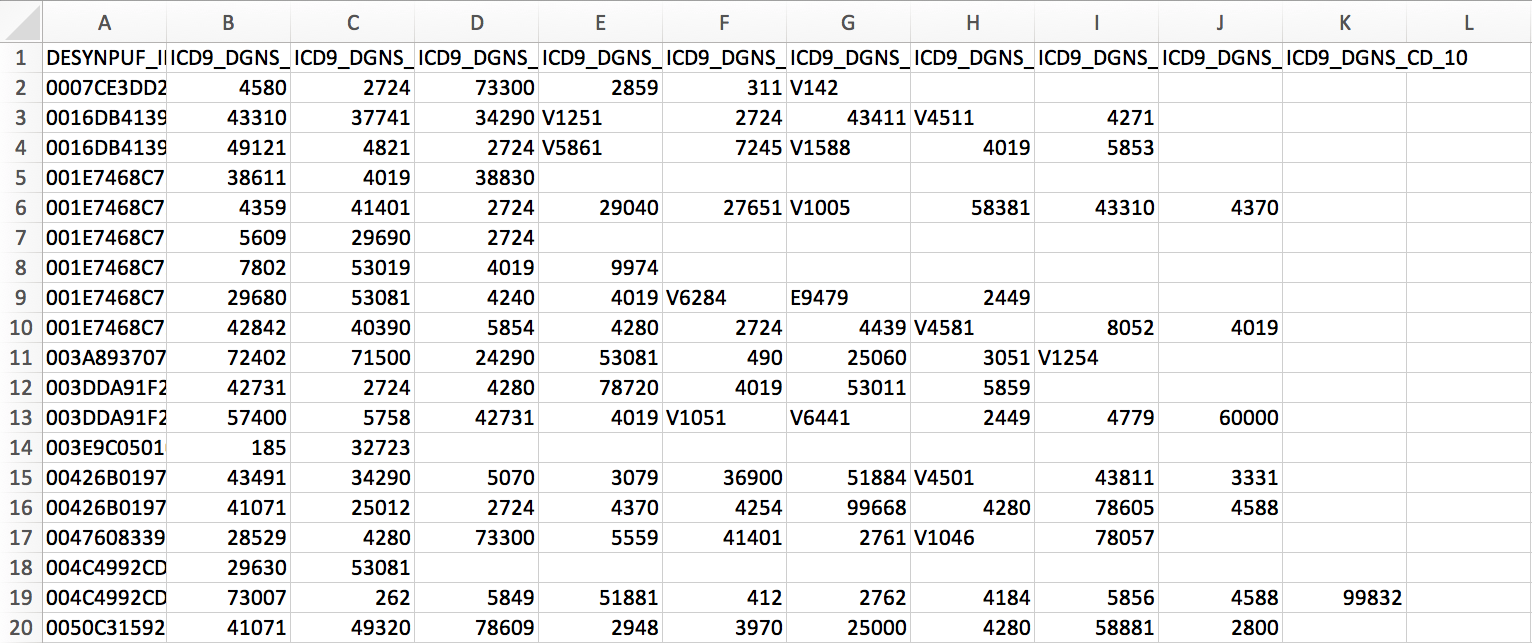
As you can see, the dataset contains 10 columns of ICD-9 codes. For example, the first patient had “4580”, “2724”, “73300”, “2859”, “311”, “V142”. By looking up the ICD-9, you will be able to translate each code to the specific disease. For example, 4580 is corresponded to “Orthostatic hypotension”, and 311 refers to “Depressive disorder”.
Processing Data#
By looking closely at the dataset, you will see some procedure codes, which starts with an letter(V4511, V142, etc). We excluded these procedure codes, in which they are referring the external causes of injury, such as “road vehicle accidents”.
for i in data.columns:
data[i][data[i].str.contains('V', na=False)] = ''
data[i][data[i].str.contains('E', na=False)] = ''
data[i] = data[i].str.lstrip('0')
The next step involved transferring the ICD-9 codes into higher hierarchy. What does that mean? The initial dataset contains four or five digits ICD-9 code, in which each code corresponds to a distinct disease. Therefore, they are at the lowest hierarchy. We’d like to start from 3 digits ICD-9 code, in which each code is recognized as a disease category. For instance, 4580 refers to Orthostatic hypotension. By cutting it to 3-digits, 458 represents Hypotension that also includes five other disease related to hypotension.
# Cut to three numbers(ICD9_Code)
data_3 = data.copy()
for i in data_3.columns:
data_3[i] = data_3[i].str[0:3]
Next, we created a dictionary with all disease code, and mapped with unique integer as follow:
from gensim import corpora
dictionary = corpora.Dictionary(processed_docs)
dictionary.token2id
[output]
'285': 1,
'311': 2,
'458': 3,
'733': 4,
'272': 5,
After we created the new dictionary, we will count the number of disease for each patient. As you can see the result below, some disease are counted more than once for the same patient. This is because the patient may have multiple disease under same category. After we transferred the ICD code to higher hierarchy, the code are counted more than 1 time. [output]
[(0, 1), (1, 1), (2, 1), (3, 2), (4, 1)],
[(0, 2), (5, 1), (6, 1), (7, 1), (8, 2), (9, 1)],
[(0, 2), (5, 1), (10, 1), (11, 2), (12, 1), (13, 1)],
[(14, 2), (15, 1)],
[(0, 1), (5, 1), (8, 1), (16, 1), (17, 1), (18, 2), (19, 1), (20, 1)],
[(5, 1), (21, 2)],
[(10, 1), (22, 2), (23, 1)],
[(0, 2), (10, 1), (24, 1), (25, 2), (26, 1)]
LDA model#
Next, we are ready to train our LDA model. One of the hyper parameters is “num_topics”. We set the number of topics to be four. After we run the model, we end up with the probabilistic topic model, which is to discover the hidden structure in large archives of documents. As shown below, we ended up with four topics.
(0,
'0.099*"" + 0.054*"414" + 0.049*"250" + 0.047*"427" + 0.046*"401" + 0.034*"272" + 0.029*"715" + 0.028*"486" + 0.028*"786" + 0.022*"530"'),
(1,
'0.096*"" + 0.048*"401" + 0.044*"599" + 0.039*"296" + 0.035*"780" + 0.027*"272" + 0.027*"250" + 0.026*"295" + 0.022*"414" + 0.022*"305"'),
(2,
'0.093*"428" + 0.055*"" + 0.049*"276" + 0.043*"518" + 0.036*"584" + 0.035*"427" + 0.030*"414" + 0.029*"585" + 0.026*"491" + 0.025*"389"'),
(3,
'0.073*"" + 0.055*"276" + 0.030*"401" + 0.029*"285" + 0.027*"724" + 0.023*"780" + 0.021*"530" + 0.021*"578" + 0.019*"250" + 0.018*"789"')

So far, we already can see the hidden topics in this inpatient dataset. We got four hidden topics in which each one contains ICD disease codes.
Clustering#
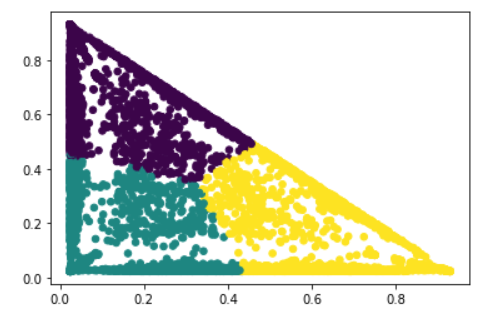
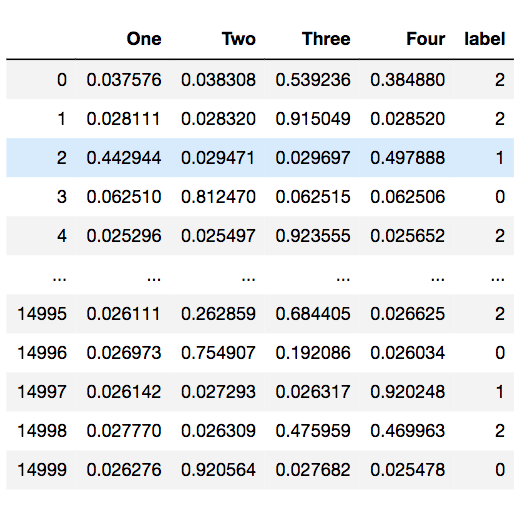
Last, we used K-means to discover the clusters based on the topic modeling result. The matrix below shows that which cluster that a patient belongs to.
 When plotting the probability of each topic, we can clearly see the three topics, which are green, blue/red, orange. (The purple area is the label name, doesn’t mean anything)
When plotting the probability of each topic, we can clearly see the three topics, which are green, blue/red, orange. (The purple area is the label name, doesn’t mean anything)
Conclusion#
For this project, we discovered hidden topics with LDA, a method of topic modeling. In healthcare, knowing this information can help professional to know the potential disease that a depressed patients will have. Lastly, we used k-means to cluster the patients into three groups. This is how LDA can be used to classify healthcare dataset.
References:#
Michael J. Paul and Mark Dredze. Factorial LDA: Sparse multi-dimensional text models. In Proceedings of the Conference on Nueral Information Processing Systems (NIPS’12), pages 2591–2599, 2012.